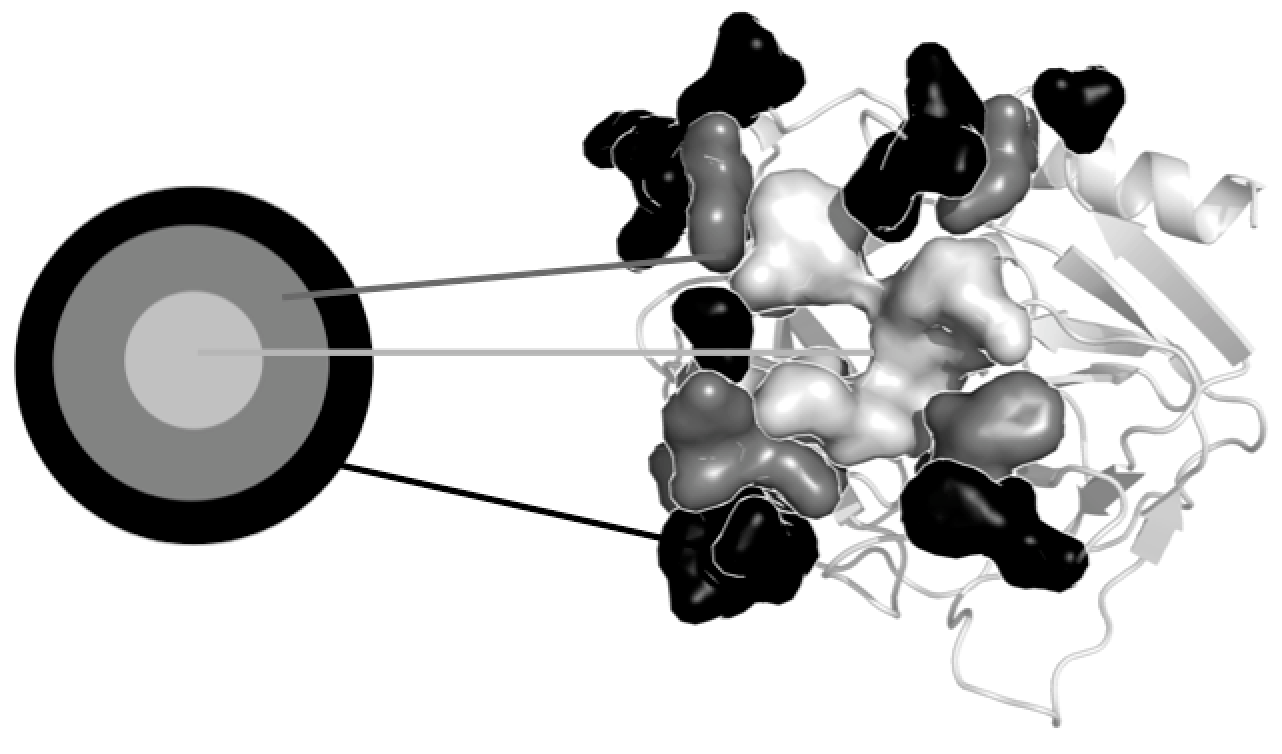
World Community Grid posts a news on Joint Evolutionary Trees 2 (JET2), a new large-scale method to predict protein-protein interfaces based on sequence and structure information.
To the Article and Software.
|
|
|
The team of Diatom Functional Genomics just published a paper describing how marine diatoms efficiently attune acclimation responses in highly variable ocean environments. L.Taddei*, G.R. Stella*, A. Rogato, B. Bailleul, A.E. Fortunato, R. Annunziata, R. Sanges, M. Thaler, B. Lepetit, J. Lavaud, M. Jaubert, G. Finazzi, J.P. Bouly, A. Falciatore. Multi-signal control of expression of the LHCX protein family in marine diatom Phaeodactylum tricornutum. Journal Experimental Botany, in press.
*co-first-authors
|
|
|
We achieved the complete reconstruction of genome history in a model yeast genus, providing a granular view of genome evolution linking gene content, chromosome rearrangements and protein divergence into a single evolutionary framework.
N Vakirlis, Sarilar V, G Drillon, A Fleiss, N Agier, J-P Meyniel, L Blanpain, A Carbone, H Devillers, K Dubois, A Gillet-Markowska, S Graziani, Nguyen H-V, M Poirel, C Reisser, J Schott, J Schacherer, I Lafontaine, B Llorente, C Neuvéglise and Fischer G. Reconstruction of ancestral chromosome architecture and gene repertoire reveals principles of genome evolution in a model yeast genus. Genome Research (2016).
|
A.E. Fortunato, M. Jaubert, J.P. Bouly, M. Thaler and A. Falciatore from the Diatom Functional Genomics team in collaboration with J.S. Bernardes (Statistical Genomics and Biological Physics), A. Carbone (Analytical Genomics) and other international collaborators published on The Plant Cell the paper “Diatom Phytochromes Reveal the Existence of Far-Red-Light-Based Sensing in the Ocean.” Full text paper.
Plant Cell Editor’s comment on this work.
|
E.Laine and A.Carbone just published a new large-scale method to predict protein-protein interfaces based on sequence and structure information - Joint Evolutionary Trees 2 (JET2). It is the first time that a method highlights, with such a precision, information on distinguished binding sites, possibly overlapping each other.
To the Article and Software.
|
|
|
|
|
|
|